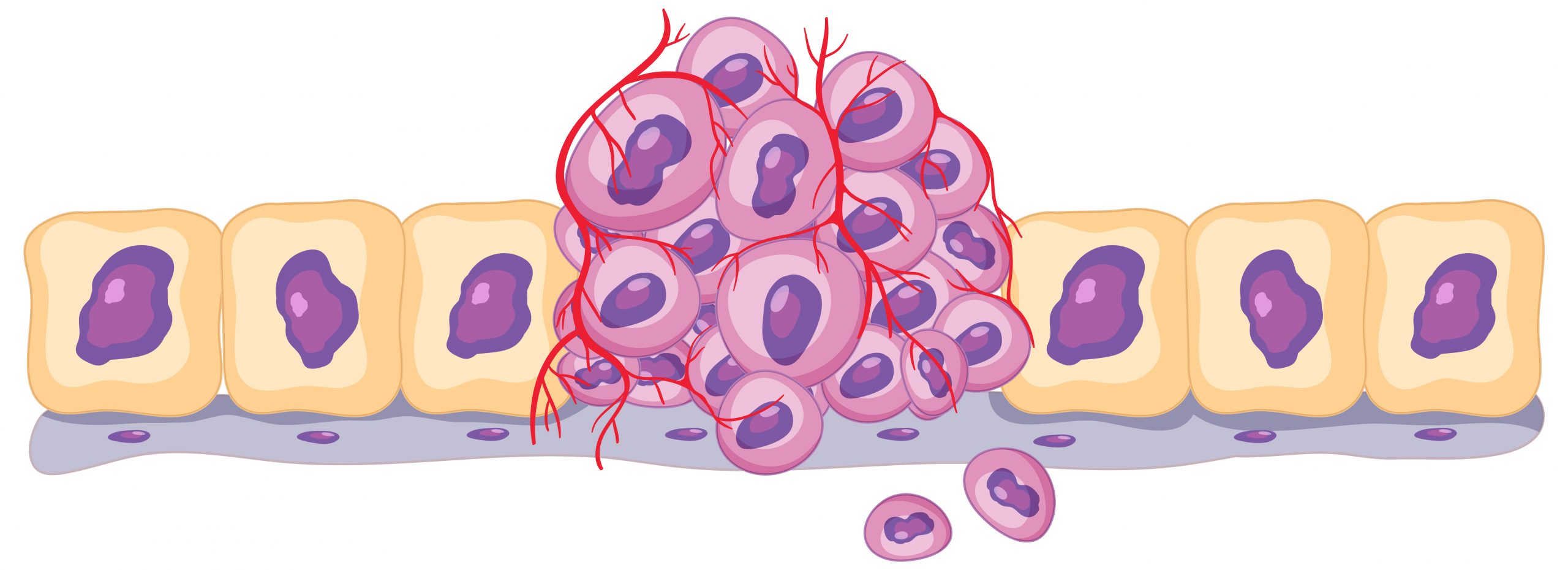

A new study led by The University of Texas MD Anderson Cancer Center researchers, published today in Nature Medicine, provides a deeper understanding of the vast diversity of T cell states, as well as their relationships and roles within the complex tumor microenvironment, bringing a new perspective to understanding immunotherapy efficacy in cancer. This new pan-cancer single-cell T cell atlas incorporates 27 single-cell RNA sequencing datasets, nine of which are unique to MD Anderson, and covers 16 cancer types. It provides the most detailed picture of the heterogeneity of T cells present within the tumor microenvironment to date, demonstrating how their phenotypic states, as well as the relative proportions of each state, play a critical role in determining the efficacy of immunotherapy and the likelihood of potential adverse effects.
“This kind of large dataset and comprehensive pan-cancer analysis provides the opportunity to see things that aren’t visible when studying a single type of cancer or even a handful of cancer types,” said corresponding author Linghua Wang, M.D., Ph.D. associate professor of Genomic Medicine. “We hope these high-resolution maps, including the thoroughly characterized T cell states, are valuable resources for facilitating future T cell studies and biomarker discovery.”
The previously unknown T cell stress response state, or TSTR, was discovered in this work. Prior single-cell investigations frequently overlooked or dismissed these T cells as aberrations of tissue separation. Using the comprehensive data available, the researchers were able to identify these cells as a distinct group, distinct from other CD4+ or CD8+ T cell subsets, and corroborate their presence in situ using multiple spatial profiling approaches.
TSTR cells might be regarded of as “stressed out” T cells, and they appear to be less effective at fighting cancer, just as a stressed person may be less effective at their job. While both TSTR cells and tired T cells are dysfunctional, TSTR cells appear to follow a distinct differentiation path from exhausted T cells.
TSTR cells have strong heat shock gene expression and, notably, are seen in much larger fractions of both CD4+ and CD8+ T cells after immune checkpoint blockade therapy, especially in non-responders. This shows that TSTR cells may play a role in immunotherapy resistance. This new T cell state adds to our understanding of cancer’s complicated biology and gives a possible target for future therapeutics.
“The fact that these TSTR cells are found in many different types of tumors opens up a whole new world of possibilities that could have high translational potential,” Wang said. Investigating the mechanistic causes of stress response in T cells, understanding how these stressed T cells are induced in the tumor microenvironment, and learning how to stop or reverse this TSTR state could catalyze the development of more effective therapeutic strategies that may bring the benefit of immunotherapy to more cancer patients.
This study further emphasizes the importance of large, integrative datasets in oncology. The capacity of big data to uncover the complicated geography of T cells within tumors is demonstrated by this pan-cancer T cell atlas. In this work, the researchers discovered seven subpopulations within the CD4+ regulatory fraction, five within the CD4+ follicular helper T cell population, and eight states among proliferating T cells.
All of these findings underscore the enormous variability of T cell states within the tumor microenvironment, as well as the need to learn more about how different states contribute to disease progression and immunotherapy response.
“There are still many questions left to answer,” Wang said. “One of the limitations of this study is we don’t have the corresponding T cell receptor data for most of the datasets analyzed. We are not sure what triggers the TSTR state, and we don’t know from which T cell subset(s) they originate. It also is unclear whether these TSTR cells are specific to tumor cells and how they communicate with and influence other cells within the tumor microenvironment.”
The T cell atlas was shared with the larger scientific community via the Single-Cell scientific platform, a user-friendly, interactive web platform. The team created this portal to allow both internal and external users to visualize and query the atlas without the need for bioinformatics expertise.
The researchers have also created a tool called TCellMap, which allows researchers to automatically label T cells from their datasets by aligning them with the high-resolution T cell maps established by this work. Wang highlighted her hope that these materials will be useful to scientists conducting in-depth analyses of T cells, leading to new discoveries and ultimately improving T cell therapy tactics.
more recommended stories
 Microglia Neuroinflammation in Binge Drinking
Microglia Neuroinflammation in Binge DrinkingKey Takeaways (Quick Summary for HCPs).
 Precision Oncology with Personalized Cancer Drug Therapy
Precision Oncology with Personalized Cancer Drug TherapyKey Takeaways UC San Diego’s I-PREDICT.
 Iron Deficiency vs Iron Overload in Parkinson’s Disease
Iron Deficiency vs Iron Overload in Parkinson’s DiseaseKey Takeaways (Quick Summary for HCPs).
 Can Ketogenic Diets Help PCOS? Meta-Analysis Insights
Can Ketogenic Diets Help PCOS? Meta-Analysis InsightsKey Takeaways (Quick Summary) A Clinical.
 Silica Nanomatrix Boosts Dendritic Cell Cancer Therapy
Silica Nanomatrix Boosts Dendritic Cell Cancer TherapyKey Points Summary Researchers developed a.
 Vagus Nerve and Cardiac Aging: New Heart Study
Vagus Nerve and Cardiac Aging: New Heart StudyKey Takeaways for Healthcare Professionals Preserving.
 Cognitive Distraction From Conversation While Driving
Cognitive Distraction From Conversation While DrivingKey Takeaways (Quick Summary) Talking, not.
 Fat-Regulating Enzyme Offers New Target for Obesity
Fat-Regulating Enzyme Offers New Target for ObesityKey Highlights (Quick Summary) Researchers identified.
 Spatial Computing Explains How Brain Organizes Cognition
Spatial Computing Explains How Brain Organizes CognitionKey Takeaways (Quick Summary) MIT researchers.
 Gestational Diabetes Risk Identified by Blood Metabolites
Gestational Diabetes Risk Identified by Blood MetabolitesKey Takeaways (Quick Summary for Clinicians).

Leave a Comment