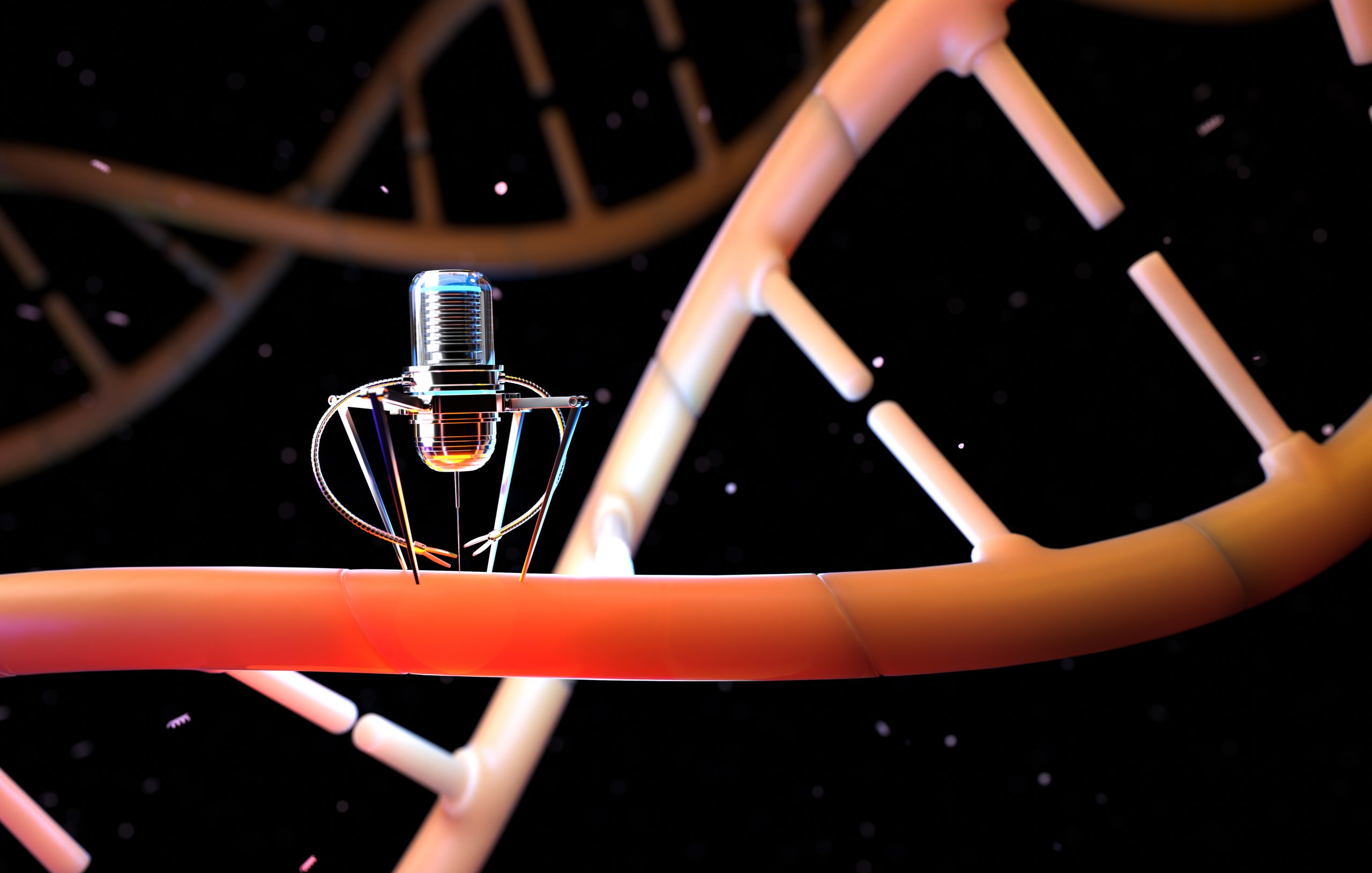

A team of scientists led by the University of Oxford has made substantial progress in identifying changes in protein structures. The method, which was reported in Nature Nanotechnology, uses novel nanopore technology to detect structural differences at the single-molecule level, even deep within lengthy protein chains.
There are roughly 20,000 protein-encoding genes in human cells. The actual number of proteins found in cells, however, is much higher, with over 1,000,000 distinct structures known. These variants are produced through a process known as post-translational modification (PTM), which takes place after a protein has been transcribed from DNA. PTM introduces structural alterations to individual amino acids that make up proteins, such as the inclusion of chemical groups or carbohydrate chains. As a result, the same protein chain can have hundreds of different variants.
These variants are important in biology because they allow for the exact regulation of complicated biological processes within individual cells. Mapping this variation would reveal a plethora of useful information that could transform our understanding of cellular processes. However, the ability to generate comprehensive protein inventories has proved elusive to date.
To address this, a team led by academics from the Department of Chemistry at the University of Oxford successfully developed a method for protein analysis based on nanopore-based sensing technology. A directional flow of water catches and unfolds 3D proteins into linear chains that are fed through microscopic pores barely wide enough for a single amino acid molecule to pass through in this method. Changes in an electrical current supplied across the nanopore are used to identify structural differences. Different molecules interrupt the current in different ways, giving them a distinct imprint.
The researchers successfully proved the method’s ability to detect three separate PTM modifications (phosphorylation, glutathionylation, and glycosylation) at the single-molecule level for protein chains longer than 1,200 residues. These comprised changes that were made deep inside the protein’s sequence. Importantly, no labels, enzymes, or other chemicals are required for the procedure.
The new protein characterisation method, according to the researchers, may be easily integrated into existing portable nanopore sequencing instruments, allowing researchers to swiftly create protein inventories of single cells and tissues. This could help with point-of-care diagnostics by allowing for the tailored detection of certain protein variations linked to diseases such as cancer and neurological disorders.
Professor Yujia Qing (Department of Chemistry, University of Oxford), contributing author for the study, said: ‘This simple yet powerful method opens up numerous possibilities. Initially, it allows for the examination of individual proteins, such as those involved in specific diseases. In the longer term, the method holds the potential to create extended inventories of protein variants within cells, unlocking deeper insights into cellular processes and disease mechanisms.’
Professor Hagan Bayley (Department of Chemistry, University of Oxford), contributing author added: ‘The ability to pinpoint and identify post-translational modifications and other protein variations at the single-molecule level holds immense promise for advancing our understanding of cellular functions and molecular interactions. It may also open new avenues for personalised medicine, diagnostics, and therapeutic interventions.’
Finally, the researchers want to transform their technology into a compact portable protein analysis device comparable to those produced by Oxford Nanopore Technologies to sequence nucleic acids (DNA and RNA). Oxford Nanopore Technologies, founded in 2005 as a spin-off from Professor Bayley’s research, has established itself as a leader in next-generation sequencing technologies. In contrast to traditional sequencing, which often needs dedicated laboratories, their unique nanopore-based approach allows scientists to sequence samples rapidly and easily utilizing available technologies. Oxford Nanopore devices have transformed fundamental and clinical genomics and played a vital role in cancer, human genetics, and infectious disease studies.
The paper titled ‘Enzyme-less nanopore detection of post-translational alterations within lengthy polypeptides’ was published in Nature Nanotechnology.
This work was done in partnership with mechanobiologist Sergi Garcia-Maynes’ research group at King’s College London and the Francis Crick Institute.
more recommended stories
 AI Predicts Chronic GVHD Risk After Stem Cell Transplant
AI Predicts Chronic GVHD Risk After Stem Cell TransplantKey Takeaways A new AI-driven tool,.
 Red Meat Consumption Linked to Higher Diabetes Odds
Red Meat Consumption Linked to Higher Diabetes OddsKey Takeaways Higher intake of total,.
 Pediatric Crohn’s Disease Microbial Signature Identified
Pediatric Crohn’s Disease Microbial Signature IdentifiedKey Points at a Glance NYU.
 Nanovaccine Design Boosts Immune Attack on HPV Tumors
Nanovaccine Design Boosts Immune Attack on HPV TumorsKey Highlights Reconfiguring peptide orientation significantly.
 High-Fat Diets Cause Damage to Metabolic Health
High-Fat Diets Cause Damage to Metabolic HealthKey Points Takeaways High-fat and ketogenic.
 Acute Ischemic Stroke: New Evidence for Neuroprotection
Acute Ischemic Stroke: New Evidence for NeuroprotectionKey Highlights A Phase III clinical.
 Statins Rarely Cause Side Effects, Large Trials Show
Statins Rarely Cause Side Effects, Large Trials ShowKey Points at a Glance Large.
 Anxiety Reduction and Emotional Support on Social Media
Anxiety Reduction and Emotional Support on Social MediaKey Summary Anxiety commonly begins in.
 Liquid Biopsy Measures Epigenetic Instability in Cancer
Liquid Biopsy Measures Epigenetic Instability in CancerKey Takeaways Johns Hopkins researchers developed.
 Human Antibody Drug Response Prediction Gets an Upgrade
Human Antibody Drug Response Prediction Gets an UpgradeKey Takeaways A new humanized antibody.

Leave a Comment